CURRENT RESEARCH
Deciphering immune recognition code at scale
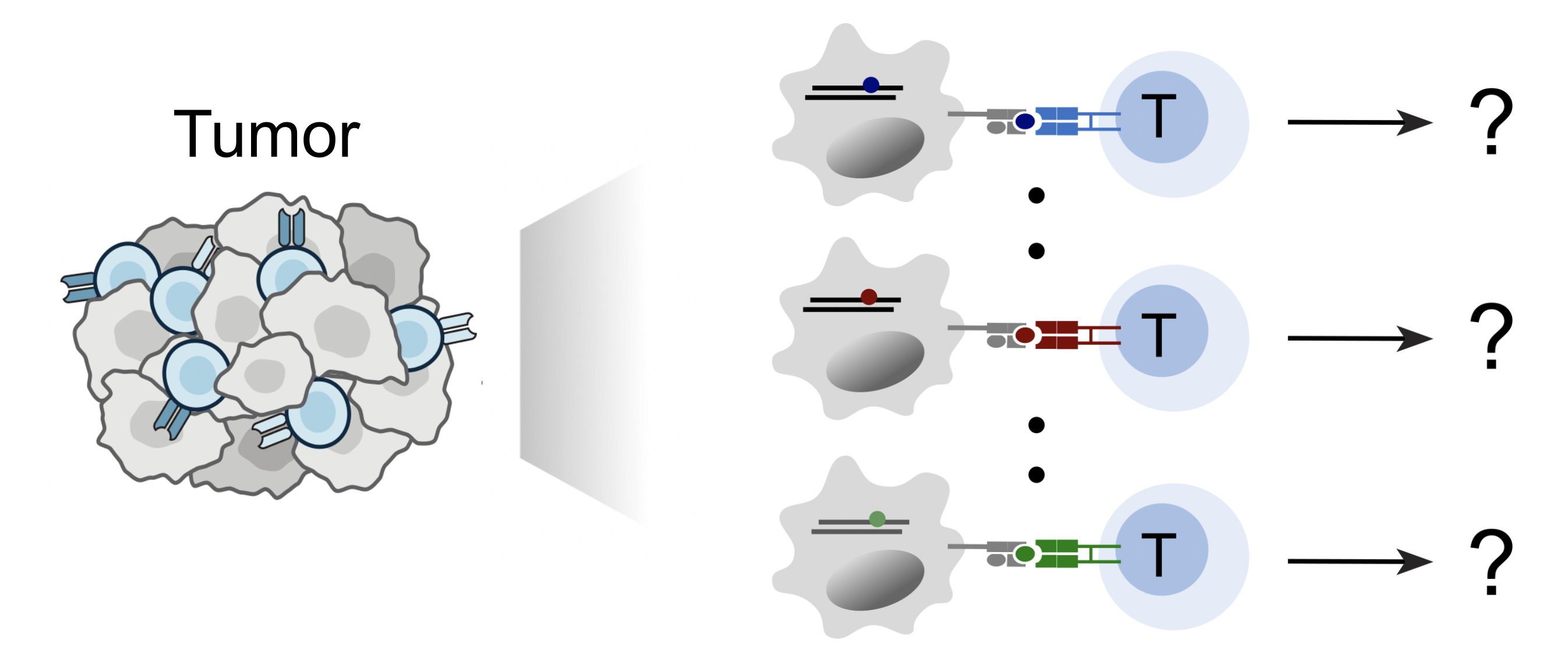
Our lab is focused on deciphering the immune recognition code, the fundamental process by which killer T cells "see" and distinguish abnormal cells from healthy ones. This is critical for the immune system to specifically target and eliminate threats like cancer cells or pathogen- infected cells. Every nucleated somatic cell presents short peptides from its entire proteome on cell surface using Major Histocompatibility Complex (MHC) molecules. T cells, equipped with a vast repertoire of T cell receptors (TCRs), scan these peptide-MHC complexes to recognize specific antigens. By integrating molecular/cellular engineering, high-throughput screening, and single-cell genomics, we develop technologies including ESCAPE-seq and ENTER-seq to tackle the following questions:
- How can we identify the specific antigens presented on MHC molecules that arise from diverse genomic alterations?
- How can we identify the T cell receptors that specifically target these MHC-presented antigens?
- How does the antigen recognition influence the subsequent fate and function of the T cells?
Exploring X-inactivation maintenance in female-biased autoimmunity
Females, harboring two X chromosomes, exhibit a heightened immune response to infections, however are more prone to develop autoimmune diseases compared to males with one X chromosome. Our recent work unveiled a critical role of female-specific lncRNA XIST in maintaining X-inactivation of key immune genes in B cells. We will further explore the molecular mechanism of X-inactivation maintenance in age-associated B cells, a subset that is abnormally expanded in patients with female-biased autoimmune diseases.
We are grateful for the funding support from sponsors who believe and invest in our research

